Fixes#92000
The documentation at https://pytorch.org/docs/stable/generated/torch.nn.MultiLabelSoftMarginLoss.html#multilabelsoftmarginloss states:
> label targets padded by -1 ensuring same shape as the input.
However, the shape of input and target tensor are compared, and an exception is raised if they differ in either dimension 0 or 1. Meaning the label targets are never padded. See the code snippet below and the resulting output. The documentation is therefore adjusted to:
> label targets must have the same shape as the input.
```
import torch
import torch.nn as nn
# Create some example data
input = torch.tensor(
[
[0.8, 0.2, -0.5],
[0.1, 0.9, 0.3],
]
)
target1 = torch.tensor(
[
[1, 0, 1],
[0, 1, 1],
[0, 1, 1],
]
)
target2 = torch.tensor(
[
[1, 0],
[0, 1],
]
)
target3 = torch.tensor(
[
[1, 0, 1],
[0, 1, 1],
]
)
loss_func = nn.MultiLabelSoftMarginLoss()
try:
loss = loss_func(input, target1).item()
except RuntimeError as e:
print('target1 ', e)
try:
loss = loss_func(input, target2).item()
except RuntimeError as e:
print('target2 ', e)
loss = loss_func(input, target3).item()
print('target3 ', loss)
```
output:
```
target1 The size of tensor a (3) must match the size of tensor b (2) at non-singleton dimension 0
target2 The size of tensor a (2) must match the size of tensor b (3) at non-singleton dimension 1
target3 0.6305370926856995
```
Pull Request resolved: https://github.com/pytorch/pytorch/pull/107817
Approved by: https://github.com/mikaylagawarecki
This PR updates the documentation for `TripletMarginLoss` in `torch.nn`. The previous version of the documentation didn't mention the parameter `eps` used for numerical stability.
This PR does the following:
1. Describes the purpose and use of the `eps` parameter in the `TripletMarginLoss` class documentation.
2. Includes `eps` in the example usage of `TripletMarginLoss`.
Please review this update for the completeness with respect to the `TripletMarginLoss` functionality. If there are any issues or further changes needed, please let me know.
Pull Request resolved: https://github.com/pytorch/pytorch/pull/105115
Approved by: https://github.com/mikaylagawarecki
About that line:
```
torch.empty(3).random_(2)
```
* Since BCE supports targets in the interval [0, 1], a better example is to sample from uniform(0, 1), using `rand`
* BCE supports multiple dimensions, and the example in `F.binary_cross_entropy` highlights it
* `rand` is more well known than `random_`, which is a bit obscure (`rand` is in the [Random Sampling section in the docs](https://pytorch.org/docs/stable/torch.html#random-sampling))
* Chaining `empty` and `random_` gives binary values as floats, which is a weird way to get that result
* Why do it in two steps when we have sampling functions that do it in a single step?
Pull Request resolved: https://github.com/pytorch/pytorch/pull/95178
Approved by: https://github.com/albanD, https://github.com/kit1980
Minor correction. `HingeEmbeddingLoss`'s documentation had this piecewise function; but there is no $\Delta$ in the function definition, it was used to denote `margin`.
$$l_n = \begin{cases}
x_n, & \text{if}\; y_n = 1,\\
\max \{0, \Delta - x_n\}, & \text{if}\; y_n = -1,
\end{cases}$$
Following other documentation guidelines, `HuberLoss` has a parameter `delta`, and its piecewise function is defined as follows; using $delta$ as a reference to the `delta` parameter and not $\Delta$.
$$l_n = \begin{cases}
0.5 (x_n - y_n)^2, & \text{if } |x_n - y_n| < delta \\
delta * (|x_n - y_n| - 0.5 * delta), & \text{otherwise }
\end{cases}$$
So by analogy, `HingeEmbeddingLoss` should also be the same, thus, the right piecewise function for it should be like the following instead.
$$l_n = \begin{cases}
x_n, & \text{if}\; y_n = 1,\\
\max \{0, margin- x_n\}, & \text{if}\; y_n = -1,
\end{cases}$$
Pull Request resolved: https://github.com/pytorch/pytorch/pull/95140
Approved by: https://github.com/albanD
This is a new version of #15648 based on the latest master branch.
Unlike the previous PR where I fixed a lot of the doctests in addition to integrating xdoctest, I'm going to reduce the scope here. I'm simply going to integrate xdoctest, and then I'm going to mark all of the failing tests as "SKIP". This will let xdoctest run on the dashboards, provide some value, and still let the dashboards pass. I'll leave fixing the doctests themselves to another PR.
In my initial commit, I do the bare minimum to get something running with failing dashboards. The few tests that I marked as skip are causing segfaults. Running xdoctest results in 293 failed, 201 passed tests. The next commits will be to disable those tests. (unfortunately I don't have a tool that will insert the `#xdoctest: +SKIP` directive over every failing test, so I'm going to do this mostly manually.)
Fixes https://github.com/pytorch/pytorch/issues/71105
@ezyang
Pull Request resolved: https://github.com/pytorch/pytorch/pull/82797
Approved by: https://github.com/ezyang
### Description
Across PyTorch's docstrings, both `callable` and `Callable` for variable types. The Callable should be capitalized as we are referring to the `Callable` type, and not the Python `callable()` function.
### Testing
There shouldn't be any testing required.
Pull Request resolved: https://github.com/pytorch/pytorch/pull/82487
Approved by: https://github.com/albanD
This PR updates the documentation for the CosineEmbeddingLoss.
The loss function uses the `cosine similarity` but in the documentation the term `cosine distance` is used. Therefor the term is changed to `cosine similarity`
Fixes#75104
Pull Request resolved: https://github.com/pytorch/pytorch/pull/75188
Approved by: https://github.com/cpuhrsch
Summary:
Changes made to line 1073: The denominator of the formula was the EXP(SUM(x)) and changed it to SUM(EXP(x))
Fixes #ISSUE_NUMBER
Pull Request resolved: https://github.com/pytorch/pytorch/pull/70220
Reviewed By: davidberard98
Differential Revision: D33279050
Pulled By: jbschlosser
fbshipit-source-id: 3e13aff5879240770e0cf2e047e7ef077784eb9c
Summary:
Pull Request resolved: https://github.com/pytorch/pytorch/pull/67443
Fixes https://github.com/pytorch/pytorch/issues/57459
After discussing the linked issue, we resolved that `F.kl_div` computes
the right thing as to be consistent with the rest of the losses in
PyTorch.
To avoid any confusion, these docs add a note discussing how the PyTorch
implementation differs from the mathematical definition and the reasons
for doing so.
These docs also add an example that may further help understanding the
intended use of this loss.
cc brianjo mruberry
Test Plan: Imported from OSS
Reviewed By: bdhirsh
Differential Revision: D32136888
Pulled By: jbschlosser
fbshipit-source-id: 1ad0a606948656b44ff7d2a701d995c75875e671
Summary:
Fixes https://github.com/pytorch/pytorch/issues/11959
Alternative approach to creating a new `CrossEntropyLossWithSoftLabels` class. This PR simply adds support for "soft targets" AKA class probabilities to the existing `CrossEntropyLoss` and `NLLLoss` classes.
Implementation is dumb and simple right now, but future work can add higher performance kernels for this case.
Pull Request resolved: https://github.com/pytorch/pytorch/pull/61044
Reviewed By: zou3519
Differential Revision: D29876894
Pulled By: jbschlosser
fbshipit-source-id: 75629abd432284e10d4640173bc1b9be3c52af00
Summary:
Fixes https://github.com/pytorch/pytorch/issues/53964. cc albanD almson
## Major changes:
- Overhauled the actual loss calculation so that the shapes are now correct (in functional.py)
- added the missing doc in nn.functional.rst
## Minor changes (in functional.py):
- I removed the previous check on whether input and target were the same shape. This is to allow for broadcasting, say when you have 10 predictions that all have the same target.
- I added some comments to explain each shape check in detail. Let me know if these should be shortened/cut.
Screenshots of updated docs attached.
Let me know what you think, thanks!
## Edit: Description of change of behaviour (affecting BC):
The backwards-compatibility is only affected for the `reduction='none'` mode. This was the source of the bug. For tensors with size (N, D), the old returned loss had size (N), as incorrect summation was happening. It will now have size (N, D) as expected.
### Example
Define input tensors, all with size (2, 3).
`input = torch.tensor([[0., 1., 3.], [2., 4., 0.]], requires_grad=True)`
`target = torch.tensor([[1., 4., 2.], [-1., 2., 3.]])`
`var = 2*torch.ones(size=(2, 3), requires_grad=True)`
Initialise loss with reduction mode 'none'. We expect the returned loss to have the same size as the input tensors, (2, 3).
`loss = torch.nn.GaussianNLLLoss(reduction='none')`
Old behaviour:
`print(loss(input, target, var)) `
`# Gives tensor([3.7897, 6.5397], grad_fn=<MulBackward0>. This has size (2).`
New behaviour:
`print(loss(input, target, var)) `
`# Gives tensor([[0.5966, 2.5966, 0.5966], [2.5966, 1.3466, 2.5966]], grad_fn=<MulBackward0>)`
`# This has the expected size, (2, 3).`
To recover the old behaviour, sum along all dimensions except for the 0th:
`print(loss(input, target, var).sum(dim=1))`
`# Gives tensor([3.7897, 6.5397], grad_fn=<SumBackward1>.`
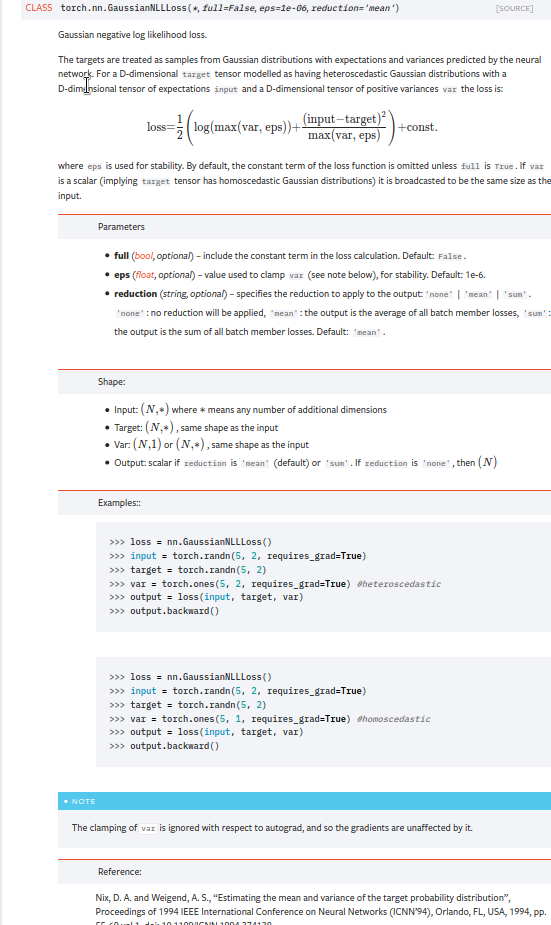
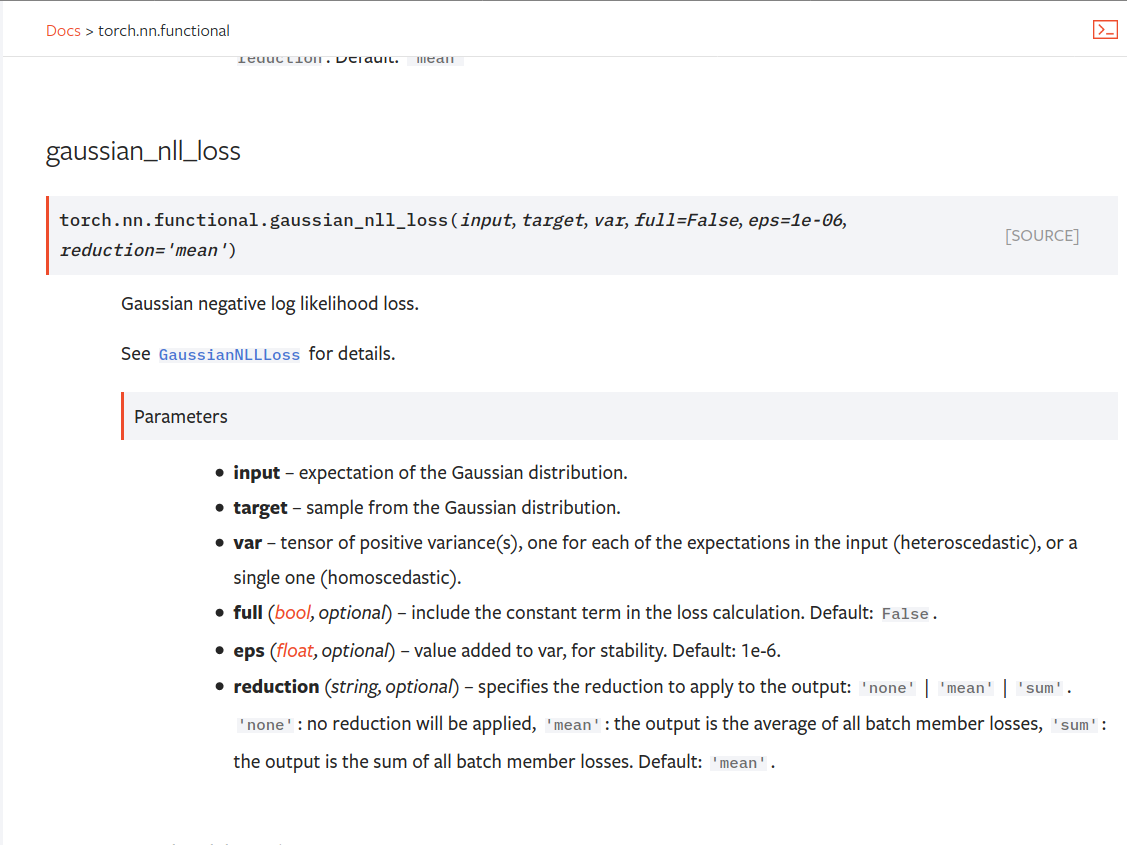
Pull Request resolved: https://github.com/pytorch/pytorch/pull/56469
Reviewed By: jbschlosser, agolynski
Differential Revision: D27894170
Pulled By: albanD
fbshipit-source-id: 197890189c97c22109491c47f469336b5b03a23f